In the current pharmaceutical industry, enzyme-catalyzed REDOX reactions play a crucial role. As the most commonly used hydrogen donors, nicotinamide adenine dinucleotide (NADH) and nicotinamide adenine dinucleotide phosphate (NADPH) account for almost 90% of biocatalytic REDOX processes.
However, wild nicotinamide adenine dinucleotide dehydrogenase (FDH) is mostly dependent on NAD and has poor catalytic activity compared to other enzymatic tools such as glucose dehydrogenase. Therefore, in order to achieve a reliable and robust NADPH regeneration system, it is necessary to develop FDH that is efficient and capable of utilizing NADP.
Jianhe Xu and his team from East China University of Science and Technology recently published a research paper entitled "Engineering a Formate Dehydrogenase for NADPH Regeneration" in the journal ChemBioChem. A new formate dehydrogenase (CdFDH) from Candida duni is reported. Through rational design guided by structure, CdFDH does not show strict NAD preference, and has good NADP utilization ability, which can support different NADph-dependent biocatalysis.

In a previous study, the team successfully engineered a NADP-dependent mutant (BstFDH-G146M/A287G) from Burkholderia stabilis through rational design.
However, further random mutations failed to produce higher catalytic activity mutants. So the researchers turned to other FDHs in search of a better starting point for protein engineering. As a new member of NADP-dependent FDH, CdFDH has become an ideal research object.
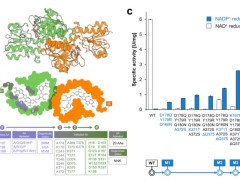
Through structure to guide the rational/half a rational design, CdFDH did not show the strict NAD dependency, and in the construction of the combination of the mutant CdFDH - M4 (D197Q/Y198R/Q199N as/A372S/K371T/delta Q375 / K167R/H16L/K159R), The catalytic efficiency of NADP (kcat/Km) was increased by 75 times.
CdFDH-M4 performs well in a variety of asymmetric REDOX processes, with its total coenzyme turnover (TTN) ranging from 135 to 986, giving it potential for use in biocatalactic reactions requiring NADPH.

The team then successfully obtained the optimal co-crystallization of CdFDH-M4 with NADP (PDB: 8J3P) at A resolution of 2.3 A.
The study found that the binding environment of nicotinamide coenzyme has changed dramatically compared to the wild type, thus leading to better NADP adaptability.
Mutation sites D197Q, Y198R, and Q199N, CdFDH-M4 provide a larger space near the hydroxyl group of NAD(H) ribose to accommodate the phosphate group, and mutation sites Gln197, Arg198, and Asn199 residues also form a hydrogen bond network with the phosphate group of NAD(P)H.
In addition, the mutation site A372S introduces an entirely new hydrogen bond network (Figure 2), and Lys3 and NAD(P)H phosphate groups from another subunit support additional polar contacts that are not observed in wild-type enzymes.

In conclusion, this study successfully modified a highly efficient NADP-dependent FDH mutant (CdFDH-M4), providing strong support for the regeneration of niacinamide coenzyme.
CdFDH-M4 has good NADP utilization ability and can support different types of NADPH-dependent biocatalactic reactions.
Structural analysis and molecular dynamics studies revealed the key role of mutation sites in promoting substrate/coenzyme binding and FDH catalyzed cyclic transformation.
At the same time, the study also provides a valuable example for expanding NADPH regeneration system, and provides inspiration for improving the cofactor preference of this FDH superfamily.